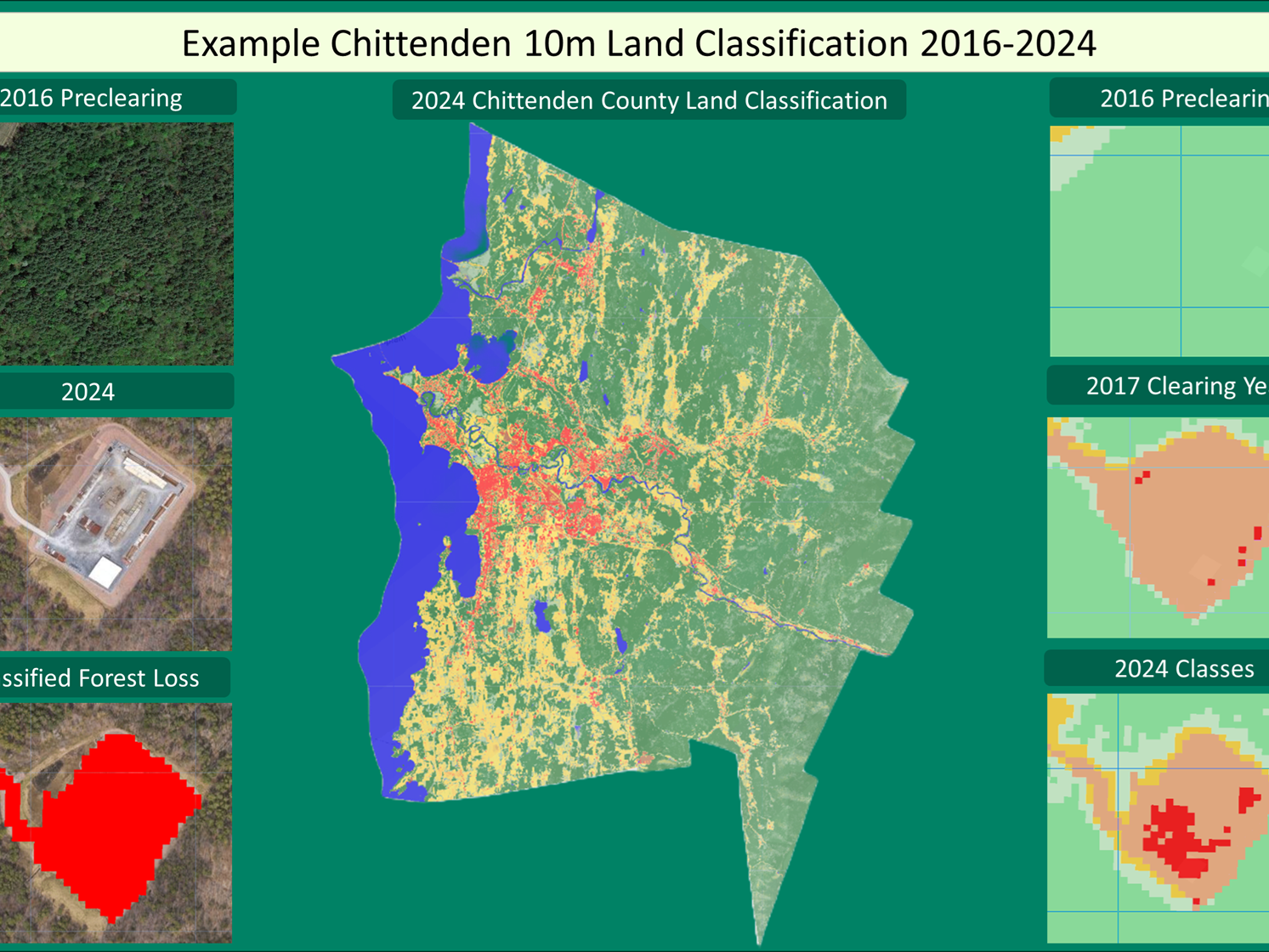
This project is centered on the analysis of recreational use, forest health, and environmental impacts within the Northeastern U.S., using various geospatial layers that combine data from OpenStreetMap (OSM), Strava, iNaturalist, and NRCS Web Soil Survey. The primary aim of the project was to identify recreational hotspots, assess the suitability of soils for sustaining such activities, and evaluate forest canopy health based on NDVI deviations from long-term baselines.
The products generated from this project include comprehensive analyses of recreational trail use, soil suitability, and wildlife impact. By integrating user-generated activity data (such as Strava and iNaturalist) with spatial environmental data (like NRCS soils and NDVI), the project provides insights into areas where recreational activity may pose risks to forest ecosystems or where management actions could be prioritized.
These layers are designed to be publicly available, ensuring that land managers, conservationists, and other stakeholders can utilize them to inform sustainable trail management, conservation efforts, and recreation planning. The datasets provide valuable tools for understanding the balance between recreational use and environmental stewardship, especially in regions where trails intersect with sensitive soils or wildlife habitats.
Results
Identified key recreational hotspots across the Northeast, including both expected and lesser-known areas of high use.
Revealed sensitive soils under high recreational pressure, particularly in major mountain ranges, indicating areas needing management intervention.
Assessed forest canopy health using NDVI data, showing that recreation may contribute to subtle impacts but larger drivers like climate remain more significant.
Highlighted trail interactions with wildlife habitats, offering data to support trail rerouting or decommissioning to enhance habitat connectivity.
Created publicly available tools for land managers to balance recreational use with ecosystem conservation.





Highlighted Methodology and Applicable Coding for NDVI Deviance from Norm
# Task 5: Create min and max rasters
def create_min_max_rasters_with_rasterio(raster_files, output_directory, excel_data):
min_array = np.inf * np.ones_like(rasterio.open(raster_files[0]).read(1), dtype=float)
max_array = -np.inf * np.ones_like(min_array)
"""
Title: ForWarn Data Processing
Author: Soren Donisvitch
Date: 11/10/2023
Dependents: Python >3, geopandas, rasterio, correct directory structure
Description: These functions provide processing and viz for processing of ForWarn derived products
Foreword: The use or application of these code without permission of the author is prohibited.The author is not liable for the use, modification, or any other application of this or other provided scripts.
"""
import os
import glob
import pandas as pd
from osgeo import gdal
from osgeo import gdal_array
import numpy as np
import matplotlib.pyplot as plt
import rasterio
import imageio
Title: ForWarn Data Processing
Author: Soren Donisvitch
Date: 11/10/2023
Dependents: Python >3, geopandas, rasterio, correct directory structure
Description: These functions provide processing and viz for processing of ForWarn derived products
Foreword: The use or application of these code without permission of the author is prohibited.The author is not liable for the use, modification, or any other application of this or other provided scripts.
"""
import os
import glob
import pandas as pd
from osgeo import gdal
from osgeo import gdal_array
import numpy as np
import matplotlib.pyplot as plt
import rasterio
import imageio
# Task 1: Create a list of all raster files in a provided directory
def get_raster_files(directory):
raster_files = glob.glob(os.path.join(directory, '*.tif'))
return raster_files
def get_raster_files(directory):
raster_files = glob.glob(os.path.join(directory, '*.tif'))
return raster_files
# Task 2: Import an Excel table with two columns (DN and pctNDVIc)
def import_excel_table(excel_path):
excel_data = pd.read_excel(excel_path)
return excel_data
def import_excel_table(excel_path):
excel_data = pd.read_excel(excel_path)
return excel_data
# Task 3: Create new rasters with pctNDVIc values instead of DN
def replace_dn_with_pctndvic(raster_files, excel_data, output_directory):
for raster_file in raster_files:
ds = gdal.Open(raster_file, gdal.GA_ReadOnly)
dn_band = ds.GetRasterBand(1)
dn_array = dn_band.ReadAsArray()
def replace_dn_with_pctndvic(raster_files, excel_data, output_directory):
for raster_file in raster_files:
ds = gdal.Open(raster_file, gdal.GA_ReadOnly)
dn_band = ds.GetRasterBand(1)
dn_array = dn_band.ReadAsArray()
# Match DN values with pctNDVIc values from the Excel table
pctndvic_array = np.zeros_like(dn_array, dtype=float)
for index, row in excel_data.iterrows():
dn_value = row['DN']
pctndvic_value = row['pctNDVIc']
pctndvic_array[dn_array == dn_value] = pctndvic_value
pctndvic_array = np.zeros_like(dn_array, dtype=float)
for index, row in excel_data.iterrows():
dn_value = row['DN']
pctndvic_value = row['pctNDVIc']
pctndvic_array[dn_array == dn_value] = pctndvic_value
# Create a new raster with pctNDVIc values
output_path = os.path.join(output_directory, f'pctndvic_{os.path.basename(raster_file)}')
write_raster(output_path, ds, pctndvic_array)
output_path = os.path.join(output_directory, f'pctndvic_{os.path.basename(raster_file)}')
write_raster(output_path, ds, pctndvic_array)
# Task 4: Make a stack of new rasters with pctNDVIc values and create a new single raster of the cumulative sum of each cell
def create_cumulative_raster_with_rasterio(raster_files, excel_data, output_directory):
with rasterio.open(raster_files[0]) as src:
result_array = src.read().astype(float)
result_array_avg = src.read().astype(float)
result_profile = src.profile
def create_cumulative_raster_with_rasterio(raster_files, excel_data, output_directory):
with rasterio.open(raster_files[0]) as src:
result_array = src.read().astype(float)
result_array_avg = src.read().astype(float)
result_profile = src.profile
for f in raster_files[1:]:
with rasterio.open(f) as src:
assert result_profile == src.profile, f'Stopping, files {raster_files[0]} and {f} do not have matching profiles'
with rasterio.open(f) as src:
assert result_profile == src.profile, f'Stopping, files {raster_files[0]} and {f} do not have matching profiles'
# Transform the new raster from DN to pctNDVIc before adding
new_array = src.read(1).astype(float)
dn_values = src.read(1)
pctndvic_values = np.zeros_like(new_array, dtype=float)
new_array = src.read(1).astype(float)
dn_values = src.read(1)
pctndvic_values = np.zeros_like(new_array, dtype=float)
for index, row in excel_data.iterrows():
dn_value = row['DN']
pctndvi_value = row['pctNDVIc']
pctndvic_values[dn_values == dn_value] = pctndvi_value
dn_value = row['DN']
pctndvi_value = row['pctNDVIc']
pctndvic_values[dn_values == dn_value] = pctndvi_value
result_array += pctndvic_values
# Ensure the result_array has the correct shape
result_array = result_array[0, :, :]
# Calculate average
result_array_avg = np.divide(result_array,len(raster_files))
result_array = result_array[0, :, :]
# Calculate average
result_array_avg = np.divide(result_array,len(raster_files))
cumulative_path = os.path.join(output_directory, 'cumulative_raster_avg.tif')
write_raster(cumulative_path, rasterio.open(raster_files[0]), result_array_avg)
write_raster(cumulative_path, rasterio.open(raster_files[0]), result_array_avg)
# Task 5: Create min and max rasters
def create_min_max_rasters_with_rasterio(raster_files, output_directory, excel_data):
min_array = np.inf * np.ones_like(rasterio.open(raster_files[0]).read(1), dtype=float)
max_array = -np.inf * np.ones_like(min_array)
for raster_file in raster_files:
with rasterio.open(raster_file) as src:
band_array = src.read(1).astype(float)
with rasterio.open(raster_file) as src:
band_array = src.read(1).astype(float)
# Transform the raster from DN to pctNDVIc
dn_values = src.read(1)
pctndvic_values = np.zeros_like(band_array, dtype=float)
dn_values = src.read(1)
pctndvic_values = np.zeros_like(band_array, dtype=float)
for index, row in excel_data.iterrows():
dn_value = row['DN']
pctndvi_value = row['pctNDVIc']
pctndvic_values[dn_values == dn_value] = pctndvi_value
dn_value = row['DN']
pctndvi_value = row['pctNDVIc']
pctndvic_values[dn_values == dn_value] = pctndvi_value
# Update min and max arrays
min_array = np.minimum(min_array, pctndvic_values)
max_array = np.maximum(max_array, pctndvic_values)
min_array = np.minimum(min_array, pctndvic_values)
max_array = np.maximum(max_array, pctndvic_values)
# Write min and max rasters
min_path = os.path.join(output_directory, 'min_raster.tif')
max_path = os.path.join(output_directory, 'max_raster.tif')
min_path = os.path.join(output_directory, 'min_raster.tif')
max_path = os.path.join(output_directory, 'max_raster.tif')
write_raster(min_path, rasterio.open(raster_files[0]), min_array)
write_raster(max_path, rasterio.open(raster_files[0]), max_array)
write_raster(max_path, rasterio.open(raster_files[0]), max_array)
# Helper function to write raster
def write_raster(output_path, src, array):
with rasterio.open(
output_path,
'w',
driver='GTiff',
width=src.width,
height=src.height,
count=1,
dtype=array.dtype,
crs=src.crs,
transform=src.transform,
) as dst:
dst.write(array, 1)
# Function to visualize a raster
def visualize_raster(raster_path, title):
ds = gdal.Open(raster_path, gdal.GA_ReadOnly)
band_array = ds.GetRasterBand(1).ReadAsArray()
def write_raster(output_path, src, array):
with rasterio.open(
output_path,
'w',
driver='GTiff',
width=src.width,
height=src.height,
count=1,
dtype=array.dtype,
crs=src.crs,
transform=src.transform,
) as dst:
dst.write(array, 1)
# Function to visualize a raster
def visualize_raster(raster_path, title):
ds = gdal.Open(raster_path, gdal.GA_ReadOnly)
band_array = ds.GetRasterBand(1).ReadAsArray()
plt.imshow(band_array, cmap='viridis')
plt.title(title)
plt.colorbar()
plt.show()
plt.title(title)
plt.colorbar()
plt.show()
# Function to create a timelapse video of pctNDVIc rasters
def create_timelapse_video(raster_files, output_path):
images = []
for raster_file in raster_files:
ds = gdal.Open(raster_file, gdal.GA_ReadOnly)
band_array = ds.GetRasterBand(1).ReadAsArray()
def create_timelapse_video(raster_files, output_path):
images = []
for raster_file in raster_files:
ds = gdal.Open(raster_file, gdal.GA_ReadOnly)
band_array = ds.GetRasterBand(1).ReadAsArray()
plt.imshow(band_array, cmap='viridis')
plt.title(f'pctNDVIc - {os.path.basename(raster_file)}')
plt.colorbar()
plt.title(f'pctNDVIc - {os.path.basename(raster_file)}')
plt.colorbar()
# Save the current figure to a temporary image file
temp_image_path = os.path.join(output_directory, 'temp_image.png')
plt.savefig(temp_image_path)
plt.close()
temp_image_path = os.path.join(output_directory, 'temp_image.png')
plt.savefig(temp_image_path)
plt.close()
# Append the image to the list for creating the timelapse video
images.append(imageio.imread(temp_image_path))
images.append(imageio.imread(temp_image_path))
# Create a timelapse video
imageio.mimsave(output_path, images, duration=1)
imageio.mimsave(output_path, images, duration=1)
# Run:
input_directory = 'forwarn/output/clipped_rasters'
excel_file_path = 'forwarn/ForWarn_Percent_NDVI_change_lut_ESRI.xlsx'
output_directory = 'forwarn/output'
input_directory = 'forwarn/output/clipped_rasters'
excel_file_path = 'forwarn/ForWarn_Percent_NDVI_change_lut_ESRI.xlsx'
output_directory = 'forwarn/output'
raster_files = get_raster_files(input_directory)
excel_data = import_excel_table(excel_file_path)
excel_data = import_excel_table(excel_file_path)
replace_dn_with_pctndvic(raster_files, excel_data, output_directory)
visualize_raster('forwarn/output/pctndvic_20210508_aoi.tif', 'pctndvic_20210508')
visualize_raster('forwarn/output/pctndvic_20210508_aoi.tif', 'pctndvic_20210508')
create_cumulative_raster_with_rasterio(raster_files,excel_data,output_directory)
create_min_max_rasters_with_rasterio(raster_files,output_directory,excel_data)
create_min_max_rasters_with_rasterio(raster_files,output_directory,excel_data)
# Visualize the cumulative raster
cumulative_raster_path = os.path.join(output_directory, 'cumulative_raster.tif')
visualize_raster(cumulative_raster_path, 'Cumulative Raster')
visualize_raster("forwarn/output/max_raster.tif", 'max')
visualize_raster("forwarn/output/min_raster.tif", 'min')
cumulative_raster_path = os.path.join(output_directory, 'cumulative_raster.tif')
visualize_raster(cumulative_raster_path, 'Cumulative Raster')
visualize_raster("forwarn/output/max_raster.tif", 'max')
visualize_raster("forwarn/output/min_raster.tif", 'min')
# Create a timelapse video of pctNDVIc rasters
timelapse_output_path = 'forwarn/output/timelapse.gif'
create_timelapse_video(raster_files, timelapse_output_path)
timelapse_output_path = 'forwarn/output/timelapse.gif'
create_timelapse_video(raster_files, timelapse_output_path)